ToxPanel for Assessing Liver and Kidney Injury
ToxPanel is a
web-based tool to assess liver and kidney injury from in vitro or in
vivo genomic data. In the field of toxicogenomics, a common
assumption is that toxicity is associated with a change in the
expression of either a single gene or a set of genes (i.e., a
module), and that a certain injury endpoint causes a specific
gene-expression response—that is, a gene signature.
Using a toxicogenomics approach, we have derived 11 liver and 8
kidney injury modules [1] from the Open Toxicogenomics
Project-Genomics Assisted Toxicity Evaluation System (TG-GATEs)
database [2]. Each injury module is uniquely associated with a
specific organ-injury phenotype (see Tables 1 and 2). TG-GATEs
contains gene-expression data from Sprague-Dawley rats exposed to
different chemicals for 4 to 29 days, with corresponding documented
histopathological injury phenotypes.
Table 1. List of liver-injury module phenotypes, grouped into general classes with the number of genes in each module.
| Inflammation | Degeneration | Proliferation | |||
| Fibrogenesis | 48 | Anisonucleosis | 65 | Bile duct proliferation | 16 |
| Cellular infiltration | 25 | Nuclear alteration | 111 | Oval cell proliferation | 126 |
| Hematopoiesis | 27 | Cytoplasmic alteration | 18 | Cellular foci | 35 |
| Single cell necrosis | 11 | Granular degeneration | 18 | ||
Table 2. List of
kidney-injury module phenotypes with the number of genes in each
module.
| Kidney injury module | |
| Necrosis | 18 |
| Fibrogenesis | 125 |
| Cellular infiltration | 42 |
| Casts (hyaline) | 23 |
| Hypertrophy | 16 |
| Degeneration | 65 |
| Dilatation | 8 |
| Inclusion bodies (cytoplasmic) | 40 |
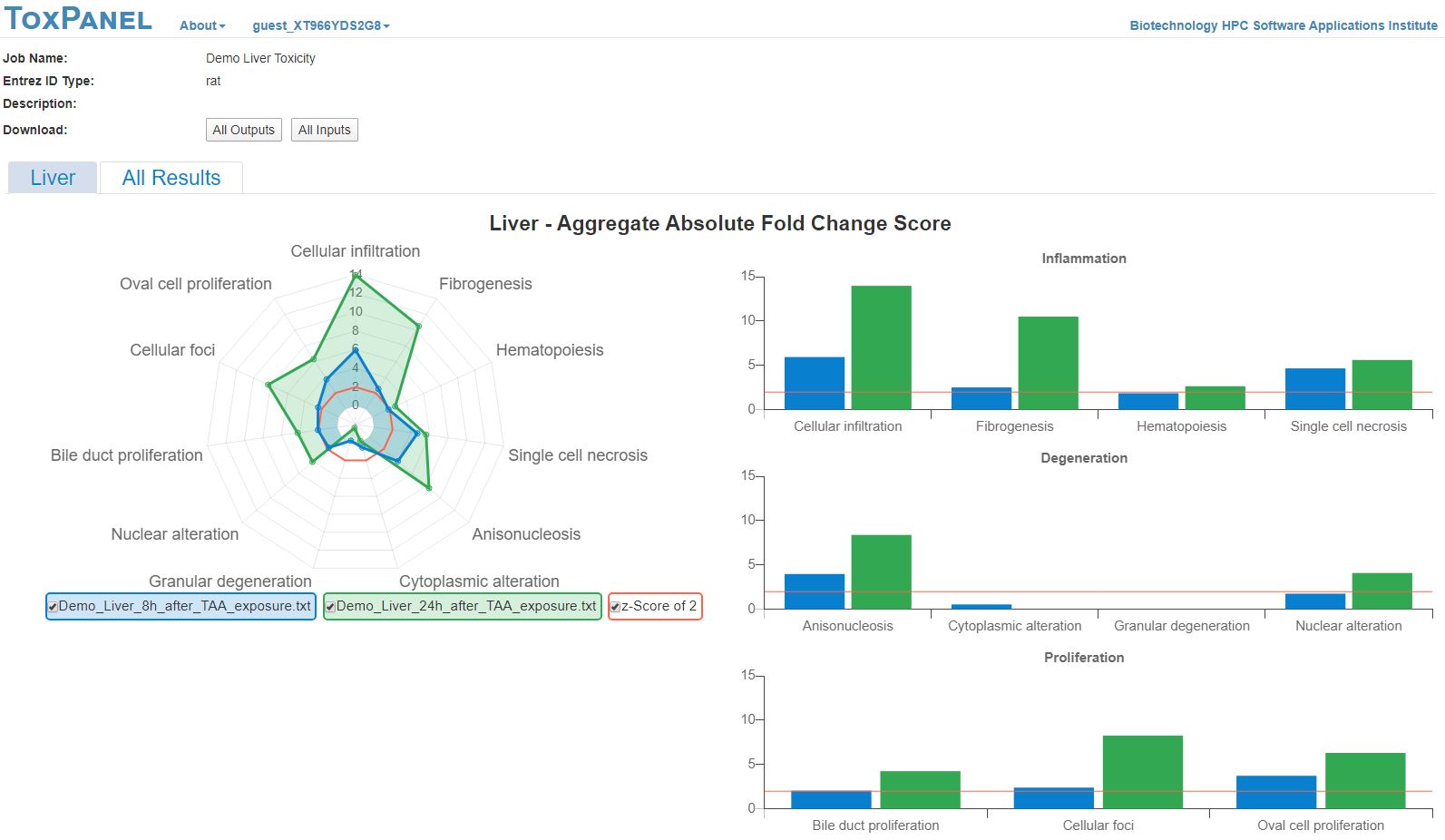
We have validated these injury modules in vivo by treating
Sprague-Dawley rats with thioacetamide [3], an organosulfur compound
extensively used in animal studies as a fibrogenesis-promoting liver
toxicant. Our
ToxPanel approach
correctly identified cellular infiltration and fibrogenesis as the
primary liver-injury phenotypes induced by thioacetamide (Figure 1).
Our liver- and kidney-injury modules can be downloaded
here.
References
- J.A. Te, M.D.M. AbdulHameed, A. Wallqvist, Systems toxicology of chemically induced liver and kidney injuries: histopathology‐associated gene co‐expression modules, J. Appl. Toxicol. 36 (2016) 1137-1149.
- Y. Igarashi, N. Nakatsu, T. Yamashita, A. Ono, Y. Ohno, T. Urushidani, H. Yamada, Open TG-GATEs: a large-scale toxicogenomics database, Nucleic Acids Res. 43 (2015) D921-D927.
- P. Schyman, R.L. Printz, S.K. Estes, K.L. Boyd, M. Shiota, A. Wallqvist, Identification of the toxicity pathways associated with thioacetamide-induced injuries in rat liver and kidney, Front. Pharmacol. 9 (2018) 1272.
- P. Schyman, R.L. Printz, S.K. Estes, T.P. O’Brien, M. Shiota, A. Wallqvist, Assessing chemical-induced liver injury in vivo from in vitro gene expression data in the rat: the case of thioacetamide toxicity, Front. Genet. 10 (2019) 1233.